High-yield DNA Extraction Kit Utilizing Magnetic Bead-based Technology that Suppresses DNA Adsorption to Soil Particles.
Extrap Soil DNA Kit Plus Ver.2
Highlights
-
DNA can be purified from a wide range of microbial species via powerful bead beating.
-
Our proprietary technology inhibits adsorption of DNA on soil particles, and it enables high-yield DNA extraction from environmental samples.
-
Easy-to-use kit with magnetic beads and applicable for automation.
-
Organic solvents such as phenol or chloroform are not required.
Product Information
Extrap Soil DNA Kit Plus ver.2 is optimized for extraction and purification of microbial DNA from environmental samples such as soil and activated sludge. Purified DNA is suitable for use in downstream applications such as microbial community structure analysis in environmental samples and real-time qPCR.
Examples of DNA Extraction
DNA Extraction Example from Various Soil Samples
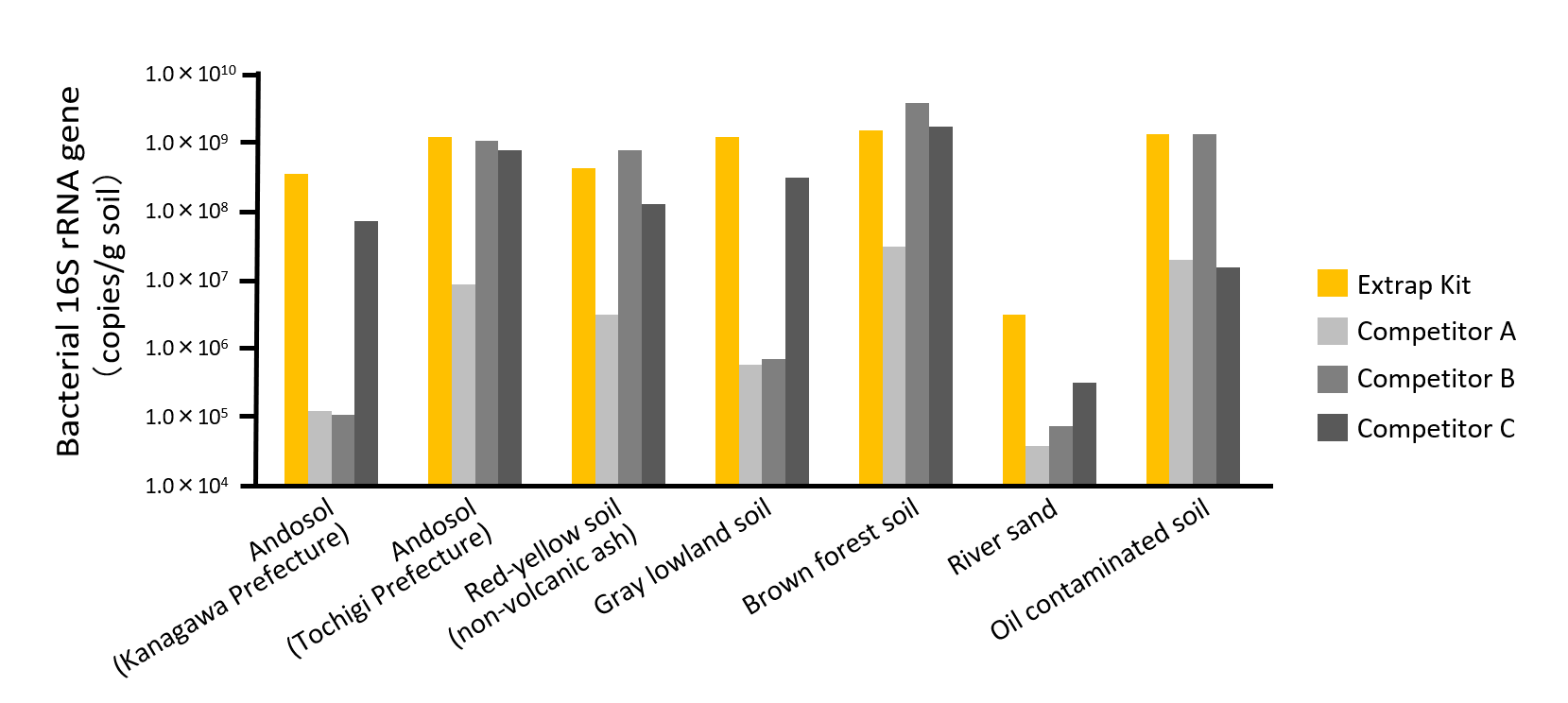
Environmental DNA was extracted from various soils which have high DNA adsorption capacity using either Extrap Soil DNA Extraction Kit or competitors’ kits. The extracted DNA was analyzed by qPCR. In all samples, Extrap Soil DNA Extraction Kit showed DNA yields that were equal to or higher than other kits.
Low Microbial DNA Contamination
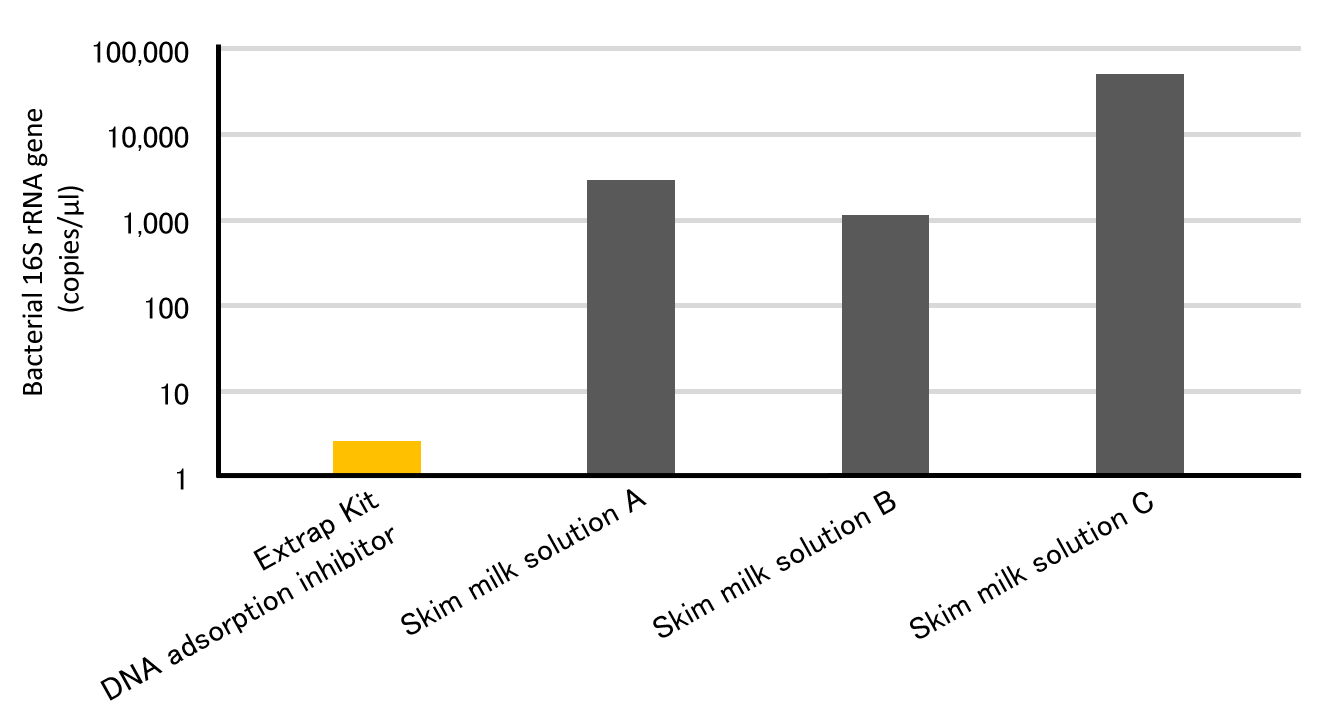
Amount of Eubacteria DNA contained in DNA adsorption inhibitors
A well-known method to prevent DNA adsorption to soil particles is addition of skim milk to DNA extraction buffer, however, it contains DNA derived from microorganisms. On the other hand, DNA adsorption inhibitor used in Extrap Soil DNA Extraction Kit contains extremely low amounts of microbial DNA. Therefore, BDL kit can be used for DNA extraction from environmental samples with low levels of microorganisms.
Procedure Overview
Sample Preparation(Beads Beating)
(1) Add environmental sample, Extraction Buffer, and Lysis Solution to a Bead Tube. Process the bead beating for 30~45 seconds.
(2) Centrifuge for 5 minutes and collect the supernatant to a 1.5 mL tube.
↓
Protein Removal
(3) Add PP Solution and mix it well. Centrifuge for 5 minutes and collect the supernatant to a 2 mL tube.
↓
DNA Purification
(4) Add MBs Solution and Binding Solution to the tube. Mix it well by inverting.
Place the tube on a magnetic stand and wait until the beads have separated from the solution. Discard the supernatant.
(5) Add Washing Solution and vortex it at low speed. Collect the magnetic beads by the magnetic stand and discard the supernatant.
(6) Add Ethanol Solution and vortex it at low speed. Collect the magnetic beads by the magnetic stand and discard the supernatant.
↓
DNA Elution
(7) After air-dry the magnetic beads, add Elution Buffer (TE buffer, sterile water, etc.).
(8) Heat at 65℃ for 10 min. Collect the magnetic beads by the magnetic stand and transfer eluted DNA solution to a new tube.
Kit Components
Bead Tubes
Extraction Buffer
Lysis Solution
PP Solution
MBs Solution
Binding Solution
Washing Solution
Citations
Ryoma K., et al., Sci. Adv., 8, (17), eabi5075 (2022).
Keiichi N., et al., Scientific Reports, 13, 2210 (2023).
Hiroyuki H., et al., Food Chemistry, 5, 100123 (2022).
Mutsumi S., et al., Chemosphere, 244, 125381 (2020).
Sandrine B., et al., Science of The Total Environment, 84, 152508 (2022).
Takashi A., et al., Science of The Total Environment, 766, 142568 (2021).
Takashi A., et al., Antibiotics , 11, (2), 210 (2022).
Akotchiffor K. G. D., et al., Mycorrhiza, 34, 119~130 (2024).
Yuki O., et al., Odontology, 112, (2), 588~600 (2024).
Ryosuke N., et al., Microbiol. Resour .Announc., 13, (3), e0127023 (2024).
Masataka A., et al., PLoS One, 18, (9), e0291742 (2023).
Daiki O., et al., BioEnergy Research, 16, 2168~2177 (2023).
Shini K., et al., Int. J. Mol. Sci., 24, (5), 4603 (2023).
Shin-Ichi H., et al., Appl. Environ. Microbiol., 89, (11), e0148823 (2023).
Takuya K., et al., Front. Ecol. Evol., 10, 959945 (2022).
Mutsumi S., et al., Biomass and Bioenergy, 164, 106551 (2022).
Wataru A., et al., Front Microbiol., 13, 1024640 (2022).
Kaho Y., et al., Water Sci. Technol., 85, (7), 2254~2264 (2022).
Zuoqian L., et al., Bioscience, Biotechnology, and Biochemistry, 86, (1), 117~124 (2021).
Miki K., et al., Bioscience, Biotechnology, and Biochemistry, 86, (12), 1705~1717 (2022).
Takashi A., et al., Antibiotics, 11, (2), 210 (2022).
Noriko T., et al., J. Water Health, 19, (4), 657~670 (2021).
Yuki I., et al., Applied and Environmental Microbiology, 10, (48), e01025-21 (2021).
Hiroki N., et al., Microbes and Environments, 36, (4), ME21029 (2021).
Yukiyo Y., et al., New Phytologist, 231, (5), 2029~2038 (2021).
Yuyao Z., et al., Water Sci. Technol., 83, (7), 1511~1521 (2021).
Rika K., et al., Microbes and Environments, 36, (2), ME20148 (2021).
Yasuko Y., et al., Microorganisms , 9, (6), 1133 (2021).
Kimiho O., et al., Extremophiles, 25, 61~76 (2021).
Kenji T., et al., PLoS One, 15, (2), e0229740 (2020).
Hiroki N., et al., Bioscience, Biotechnology, and Biochemistry, 84, (9), 1921~1935 (2020).
Shogo T., et al., Journal of Water and Environment Technology, 17, (2), 67~75 (2019).
Kimiho O., et al., Archives of Microbiology, 201, 969~982 (2019).
Kenji T., et al., Limnology and Oceanography, 64, (6), 2441~2454 (2019).
Taira H., et al., Water Sci. Technol., 80, (12), 2320~2327 (2019).
Raphaël M., et al., The ISME Journal, 14, 2907~2922 (2020). (Reference 29)
Kohei I., et al., The ISME Journal, 12, 31~47 (2018).
Jing Y., et al., Int. J. Environ. Res. Public Health, 15, (6), 1252 (2018).
Sri M., et al., asain journal of pharmaceutical and clinical research , 11, (6), 186~189 (2018).
Michio F., et al., PLoS One, 12, (12), e0189609 (2017).
Alex W., et al., The ISME Journal, 11, (8), 1915~1929 (2017).
Kentaro M., et al., Front. Bioeng. Biotechnol., 5:14 (2017).
Yasuko Y., et al., FEMS Microbiology Ecology, 91, (9), fiv093 (2015).
Kenji T., et al., Journal of Oceanography, 71, 675~683 (2015).
Yuki H., et al., Case Rep. Ophthalmol., 3, (3), 291~297 (2012).
Related Product:Magtrap
 |
|
BDL is a member of the Funakoshi Group.
 EN
EN